RESEARCH
Post-transcriptional regulation of gene expression in bacteria
Our lab investigates the coordination between RNA targeting, degradation, and translation in bacteria. Many bacteria are beneficial or harmless, but some cause infectious diseases, often regulated by RNA chaperone protein Hfq and small RNAs (sRNAs). Hfq facilitates base pairing between sRNAs and target mRNAs, leading to changes in translation initiation or RNA degradation.
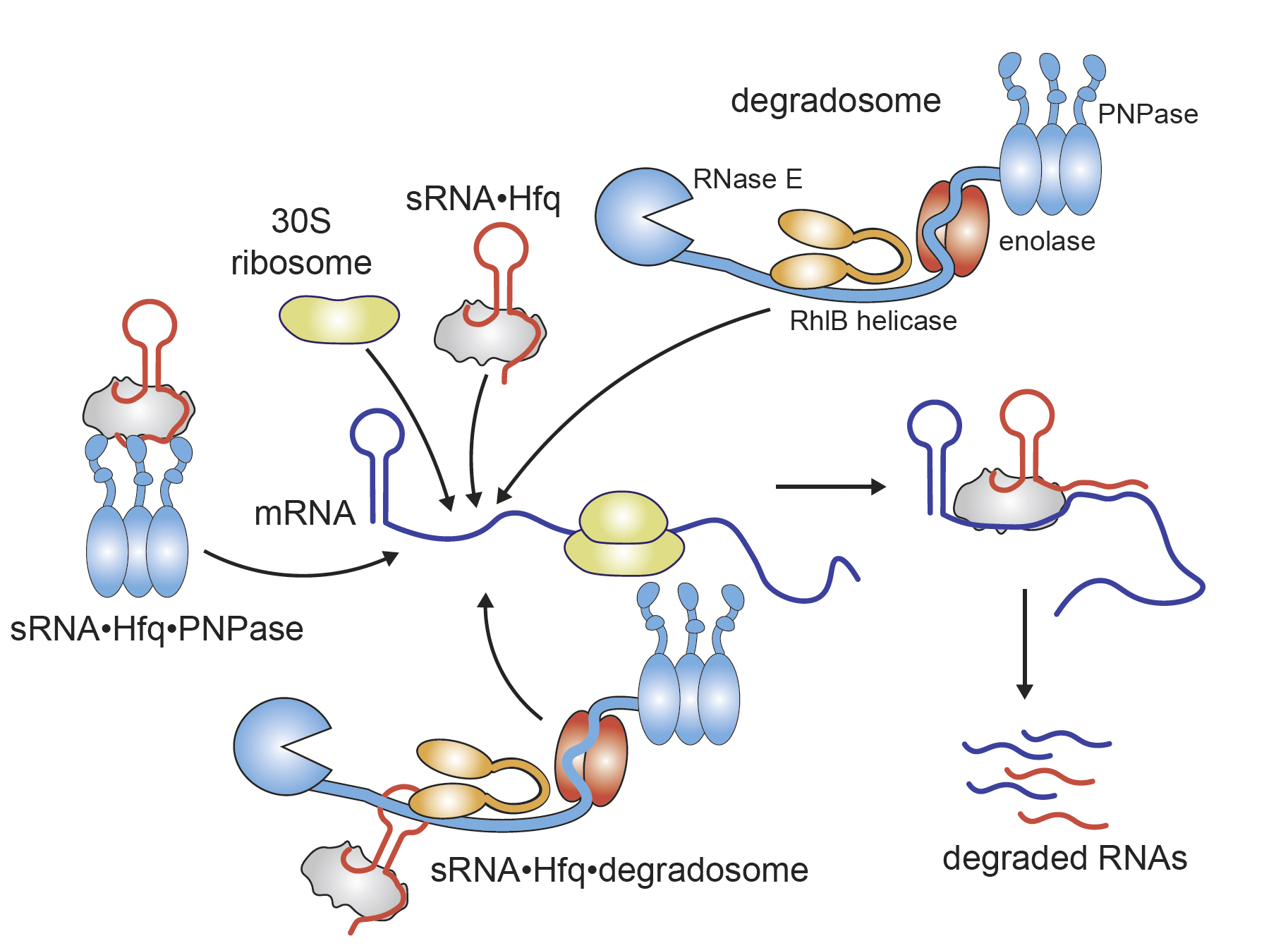
RNA degradation is carried out by the degradosome, a complex of RNase E, enolase, RhlB helicase, and PNPase. The degradosome can preassemble with sRNA and Hfq, potentially enabling more efficient regulation of mRNA. We study how alternative assembly pathways of these complexes influence regulatory outcomes.
Additionally, we aim to understand how these complexes interact with mRNA during translation. By reconstituting active translation in vitro, we investigate the real-time coordination of RNA targeting and degradation, shedding light on this critical regulatory process.
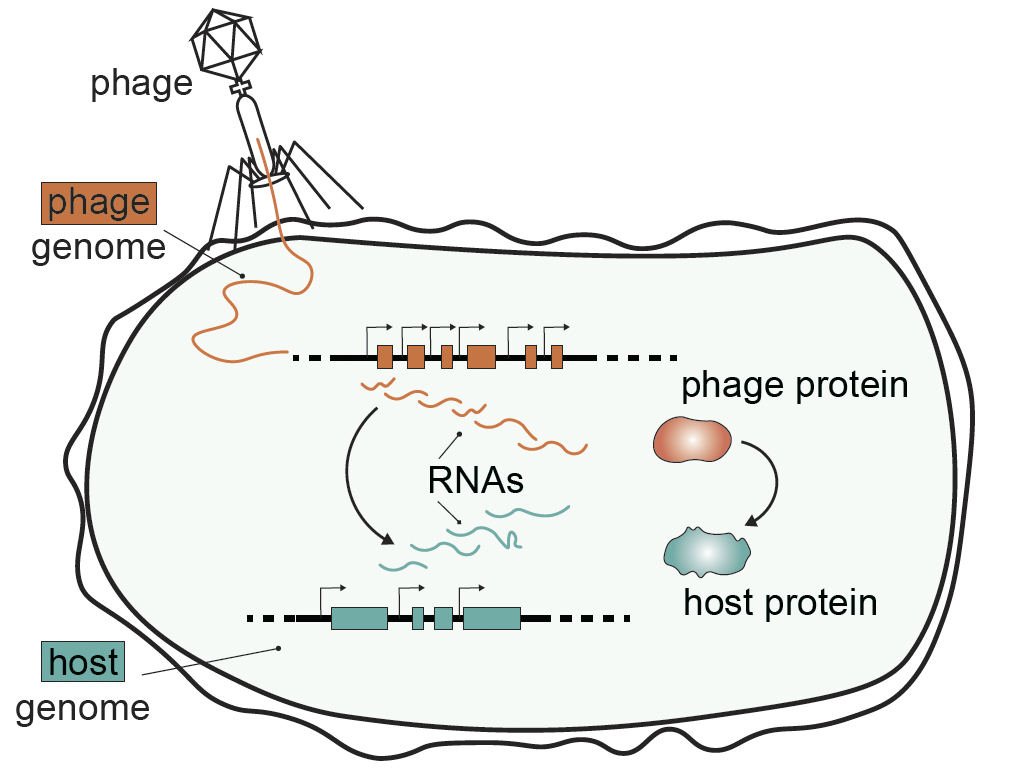
Phage-host interactions
Antibiotic resistance, driven by bacterial adaptation, poses a significant challenge, particularly with ESKAPE pathogens such as carbapenem-resistant Acinetobacter baumannii (CRAB). Bacteriophages, or phages, which are viruses that infect and lyse bacterial cells, offer a promising alternative to traditional antibiotics. Our research focuses on mapping interactions between phages and bacterial hosts across multiple biological levels to identify phage factors with antimicrobial potential. We are studying Acinetobacter and scherichia phages using transcriptomic and proteomic analyses, as well as biochemical and single-molecule approaches.
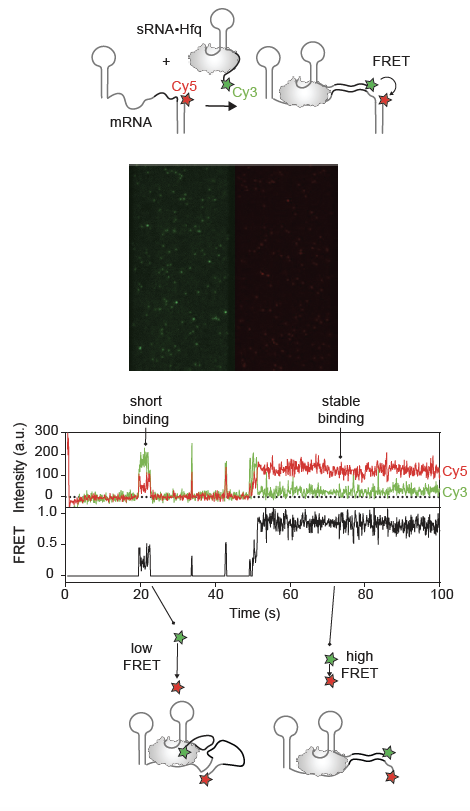
Single-molecule TIRF microscopy
Significant effects of sRNAs in gene expression are observed as quickly as 1-2 minutes from signal induction, yet we do not fully understand how the coordination between these processes occurs so efficiently on this time scale. To address this issue, we need to monitor the assembly of RNA-protein complexes as it evolves in real-time on time scales relevant in vivo. Moreover, the fate of individual sRNAs can vary, e. g. some sRNAs might fail to bind the protein or to find a cognate mRNA.
To dive deep into the molecular mechanism of fast biological processes, we need a method that:
provides a high temporal resolution (scale of milliseconds to minutes),
provides spatial resolution to resolve changes in the conformation of RNA-protein or RNA-RNA complexes,
allows the visualization of various pathways of the reaction.
Single-molecule TIRF microscopy allows the visualization of hundreds of biomolecules immobilized on the microscopic slide. However, each molecule is analyzed separately. Molecules are fluorescently labeled, and the interactions are detected by colocalization of fluorophores or FRET.